Multilevel URL Community Detection with Infomap
Posted on March 13 2017 in Machine Learning
Recently I've discovered a dataset online that is a collection of 20M web queries from about 650K users over three months from March 1 to May 31 2006.
The columns are listed as follows:
- User ID - an anonymous user ID number
- Query - the query submitted by the user
- Query Time - the time at which the query was submitted for search
- Item Rank - if the user clicked on a search result, the rank of the item on which they clicked is listed
- Click URL - if the user clicked on a search result, the domain portion of the URL in the clicked result is listed
A more detailed description of the dataset can be found here.
My goal is to find clusters of queries or url domains that are semantically and syntactically similar based on queries submitted or urls visited by a set of users. For example, a query find baby food is correlated with healthy baby food or Gerber Good Start, a brand name for baby food. Likewise, domains healthplans.com and ehealthinsurance.com can be grouped under the health insurance category. I've decided to cluster url domains, but you can always try with queries instead and compare which of the two results gives the better cluster representation.
The dataset is somewhat large, so I took careful steps to sample about 1000 users and 7500 domains after removing rows with generic, non-informative queries such as google and yahoo along with domains like google.com or - and after defining an active user to be a user with more than 10 rows. Only usr and click_url columns are used to cluster url domains. Below is the R script that preprocesses the dataset.
library(data.table)
# http://sing.stanford.edu/cs303-sp10/assignments
f <- fread("user-ct-test-collection-01.txt")
setnames(f, c('usr', 'qry', 'qry_time', 'item_rank', 'click_url'))
qdat <- f[, .N, qry][order(-N)]
rdat <- f[, .N, click_url][order(-N)]
f <- f[!click_url %in% rdat$click_url[1:100]][!qry %in% qdat$qry[1:50]]
udat <- f[, .(rows = .N,
qrys = uniqueN(qry),
urls = uniqueN(click_url)), usr][order(-rows)]
udat <- udat[rows > 10]
set.seed(54321)
udat <- udat[usr %in% sample(usr, 1000)]
set.seed(12345)
fsub2 <- f[usr %in% udat$usr][click_url %in% sample(click_url, 10000)]
fr <- unique(fsub2[, .(usr, click_url)])
uniqueN(fr$usr); uniqueN(fr$click_url) # 998 users, 7735 urls
fwrite(fr, file = "session-url-data.csv", col.names = FALSE)
Once I've sampled a set of users, create a \(N\) by \(N\) matrix of url domain similarity where \(N\) is the total number of domains and each element of the matrix is the Jaccard index between a set of users to the first domain and to the second domain. For example, let's assume that a set of users numbered 1, 2, and 3 visited the webpage healthplans.com and a set of users 1, 3, 4, and 5 visited ehealthinsurance.com. The Jaccard similarity index of the two domains is 2/5 or 0.4 since there are two overlapping users (1 and 2) out of 5 total users (1, 2, 3, 4 and 5).
Below is the Python code I wrote where the main() function computes a url domain similarity matrix, while the metric() function gives different measures of similarity between domains and the write_pajek_list() function writes the resulting matrix into a .net format. Note that I write urls when in fact I mean domains. Only the elements whose Jaccard indices are greater than 0.85 get written to the output file.
#!/usr/bin/python
from numpy import array
from collections import deque
import csv
def main():
usr_url_pairs = set()
urls = set()
users = set()
url_dict = {}
with open('session-url-data.csv', 'r') as f:
reader = csv.reader(f, delimiter = ',')
for row in reader:
usr_url_pairs.add((row[0], row[1]))
urls.add(row[1])
users.add(row[0])
if row[1] in url_dict:
url_dict[row[1]].add(row[0])
else:
url_dict[row[1]] = set(row[0])
# Convert a set into a list gives a deduped ordered list of the urls
urls = list(urls)
url_count = len(urls)
user_count = len(users)
print 'Number of URL Domains: ' + str(url_count)
print 'Number of Users: ' + str(user_count)
distance_matrix = []
link_list = deque()
# Construct a matrix of url-url similarity
i = 0
for q in urls:
distance_matrix.append(range(url_count))
j = 0
if len(distance_matrix) % 100 == 0:
print 'URL domains computed: %s' % len(distance_matrix)
while j < i:
r = urls[j]
similarity = metric(url_dict[q], url_dict[r], method = 'jaccard', total_users = user_count)
distance_matrix[i][j] = similarity
if j < i:
link_list.append((i+1, j+1, distance_matrix[i][j]))
j += 1
i += 1
write_pajek_list(link_list, urls, url_count)
def write_pajek_list(link_list, url_list, url_count):
with open('link_list.net', 'w') as f:
f.write('*Vertices ')
f.write(str(url_count))
f.write('\n')
i = 0
for row in url_list:
f.write(str(i+1) + ' "' + row + '"\n')
i += 1
f.write('*Edges ' + str(len(link_list)) + '\n')
for row in link_list:
if row[2] > .85:
f.write('%s\t%s\t%s\n' % (row[0], row[1], row[2]))
def metric(v, w, method = 'jaccard', total_users = 0):
if method == 'jaccard':
J = float(len(v & w))/len(v | w)
return J
elif method == 'normalized':
J = (float(len(v & w))/len(w))/(len(v)/float(total_users))
return J
elif method == 'distance':
J = sqrt(len(v - w) + len(w - v))
return J
else:
return 0
if __name__=='__main__':
main()
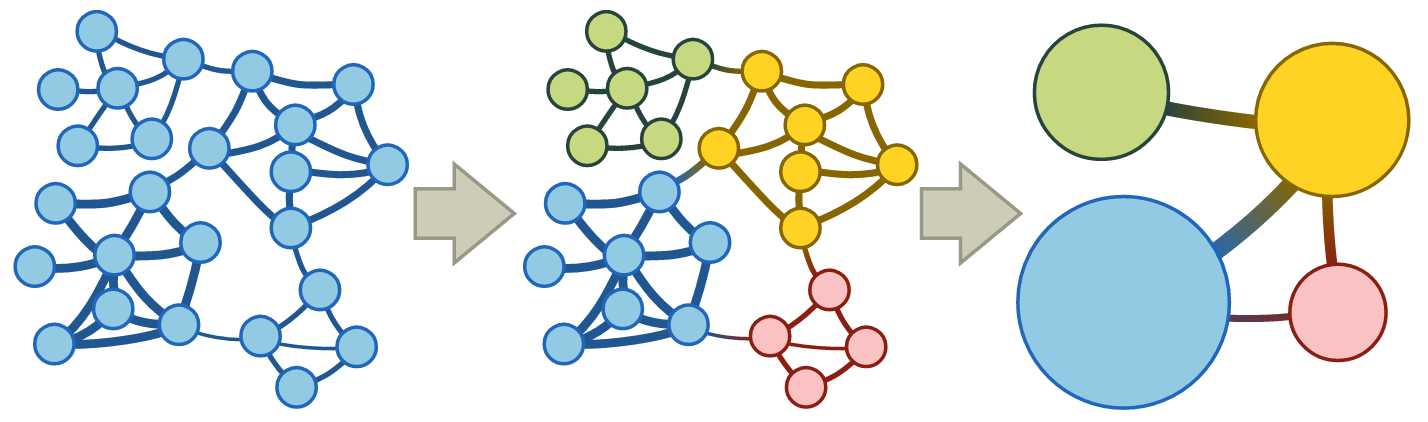
It's reasonable to convert a url domain similarity matrix into a graphical network where a node is a domain and an edge between two nodes is a degree of similarity between two domains. I've decided to use the state-of-the-art multilevel community detection with the map equation framework and its accompanying search algorithm Infomap. This paper provides a detailed explanation and tutorial of the method. Basically, the algorithm partitions a large network by detecting different communities where a set of nodes form a community when more edges are seen among nodes within the community than between other nodes not in the community. A community detection problem is inherently different from a clustering analysis, but here I use the word cluster and community interchangeably. Unlike other community detection algorithms out there, the Infomap algorithm finds communities of hierarchical structure where smaller nested communities are semantically more specific.
Once you download the source code from its main site, run the following in the folder. Feel free to tune the algorithm by adding other parameters.
./Infomap link_list.net output/ -N 10 --tree --bftree
Take a look at link_list.tree file. Note that 1:2, for example, explicitly encodes a hierarchical path in that the domain on the right is part of the second nested community within the larger first community. The clusters seem quite robust both semantically and syntactically, though they can certainly be improved. For example, it's easy to see the majority of pet food and amenities sites grouped into a single community, and the majority of real estate sites in another.
Finding clusters of similar url domains, or similar queries for that matter, is useful for online marketing by showing targeted ads to a group of users who visit certain kinds of sites more often than others. If a user is looking for cheap auto insurance and visiting sites like geico.com and progressive.com, then it's better to show display ads about auto insurance in similar sites. This kind of clustering is also useful for url domain or query expansion as semantically similar webpages or queries are not necessarily syntactically similar. For example, kimchi and Korean food might not be syntactically similar, but kimchi is a type of Korean food which results in a nested structure.